FABIA
Factor Analysis for Bicluster Acquisition (FABIA) is a model-based technique for biclustering, that is clustering rows and columns simultaneously. FABIA is a multiplicative model that assumes realistic non-Gaussian signal distributions with heavy tails. FABIA utilizes well understood model selection techniques like variational approaches and applies the Bayesian framework. The generative framework allows FABIA to determine the information content of each bicluster to separate spurious biclusters from true biclusters. On 100 simulated data sets with known true, artificially implanted biclusters, FABIA clearly outperformed all 11 competitors. FABIA was tested on microarray data sets which known, biological verfified subclusters and performed on average best out of 11 biclustering approaches.Please cite:
Sepp Hochreiter, Ulrich Bodenhofer, Martin Heusel, Andreas Mayr, Andreas Mitterecker, Adetayo Kasim, Tatsiana Khamiakova, Suzy Van Sanden, Dan Lin, Willem Talloen, Luc Bijnens, Hinrich W.H. Göhlmann, Ziv Shkedy, and Djork-Arné Clevert. FABIA: Factor Analysis for Bicluster Acquisition, Bioinformatics 2010, 26(12):1520-1527, doi:10.1093/bioinformatics/btq227
Download the R-package for Unix (source package):
- Current release:
fabia_1.4.0.tar.gz (v1.4.0 13-04-2011, build R2.13, 800 kB)
fabiaData_1.0.0.tar.gz: Demo data sets (18-05-2010, build R2.12, 5.6 MB) - Developer build:
- Previous versions:
fabia_1.3.1.tar.gz (v1.3.1 31-12-2010, build R2.13, devel, 645 kB)
fabia_1.3.1.tar.gz (v1.3.1 31-12-2010, build R2.12.1, 645 kB)
fabiaData_1.0.0.tar.gz: Demo data sets (18-05-2010, build R2.12, 5.6 MB)
fabia_1.0.0.tar.gz (v1.0.0 26-07-2010, build R2.12, 629 kB)
fabiaData_1.0.0.tar.gz: Demo data sets (18-05-2010, build R2.12, 5.6 MB)
fabia_1.0.0.tar.gz (v1.0.0 26-07-2010, build R2.11, 629 kB)
fabiaData_1.0.0.tar.gz: Demo data sets (18-05-2010, build R2.11, 5.6 MB)
fabia_1.0.0.tar.gz (v1.0.0 26-07-2010, build R2.10, 629 kB)
fabia_0.1.3.tar.gz (v0.1.3 23-04-2010, 6.2 MB)
fabia_0.1.2.tar.gz (v0.1.2 23-12-2009, 6.1 MB)
fabia_0.1.1.tar.gz (v0.1.1 28-10-2009, 5.9 MB)
Download the R-package for Windows 32-bit (i386):
- Current release:
fabia_1.4.0.zip (v1.4.0 14-04-2010, build R2.13, 1.4 MB)
fabiaData_1.0.0.zip: Demo data sets (18-05-2010, build R2.12, 5.7 MB) - Developer build:
- Previous versions:
fabia_1.3.1.zip (v1.3.1 31-12-2010, build R2.13, devel, 1.1 MB)
fabia_1.3.1.zip (v1.3.1 31-12-2010, build R2.12.1, 1.1 MB)
fabiaData_1.0.0.zip: Demo data sets (18-05-2010, build R2.12, 5.7 MB)
fabia_1.0.0.zip (v1.0.0 26-07-2010, build R2.12, 1.9 MB)
fabia_1.0.0.zip (v1.0.0 26-07-2010, build R2.11, 1.1 MB)
fabiaData_1.0.0.zip: Demo data sets (18-05-2010, build R2.11, 5.7 MB)
fabia_1.0.0.zip (v1.0.0 26-07-2010, build R2.10, 1.1 MB)
fabia_0.1.3.zip (v0.1.3 23-04-2010, 6.8 MB)
fabia_0.1.2.zip (v0.1.2 23-12-2009, 5.8 MB)
fabia_0.1.1.zip (v0.1.1 28-10-2009, 5.9 MB)
Download the R-package for Windows 64-bit (x64):
- Current release:
fabia_1.4.0.zip (v1.4.0 13-04-2010, build R2.13, 1.4 MB)
fabiaData_1.0.0.zip: Demo data sets (18-05-2010, build R2.12, 5.7 MB) - Previous versions:
fabia_1.3.1.zip (v1.3.1 31-12-2010, build R2.13, devel, 1.6 MB)
fabia_1.0.0.zip (v1.0.0 26-07-2010, build R2.12, 1.6 MB)
fabia_1.0.0.zip (v1.0.0 26-07-2010, build R2.11, 1 MB)
Paper, manuals, examples, and documents:
- FABIA paper (2.5 MB): Bioinformatics Advance Access published online on April 23, 2010
- BIOINF-2009-2000.R1-Supp.pdf (11 MB): Supplementary material
- fabia.pdf (580 kB): Software manual (vignette)
- fabia-example.pdf (21 MB): Examples of using FABIA with statistics and plots
Sources used in our experiments:
-
scripts: R chunks and tools to perform the experiments according to the paper
scripts.tar.gz (17 kB, unix)
scripts.zip (24 kB, windows)
-
scripts_supp: R chunks and tools to perform experiments M1-M3 according to the supplement
convertFABIA2biclust.R: Function to convert FABIA clusters into "biclust" class (CRAN biclust)
scripts_supp.tar.gz (7 kB, unix)
scripts_supp.zip (8 kB, windows)
- Download the R packages (unix)
truecluster_0.3.tar.gz (152 kB) contains the Munkres assignment algorithm
biclust_0.8.1.tar.gz (1 MB)
fabia_0.1.1.tar.gz (5.9 MB)
isa2_0.2.tar.gz (5.9 MB) - Download the R packages (windows)
truecluster_0.3.zip (429 kB) contains the Munkres assignment algorithm
biclust_0.8.1.zip (1.2 MB)
fabia_0.1.1.zip (5.9 MB)
Data sets and results:
- benchmark: our benchmark data set with results and evaluation
- gene_expession: three gene expression data sets with results
Copy Number Variation (CNV) Analysis with FABIA
We analyzed copy number variations (CNVs) of HapMap individuals with FABIA on the data set of (McCarroll et al. 2008), who determined CNVs of the HapMap individuals of phase II by Affymetrix Genome-Wide Human SNP Arrays 6.0.
HapMap:
The international HapMap project is a multi-country effort to identify
and catalog genetic similarities and differences in human
beings. The goal of the international HapMap project is to compare the
genetic sequences of different individuals to identify chromosomal
regions where genetic variants are shared (see http://hapmap.ncbi.nlm.nih.gov/index.html.en).
The four populations that are considered in HapMap phase II:
- Yoruba in Ibadan, Nigeria (YRI)
- Japanese in Tokyo, Japan (JPT)
- Han Chinese in Beijing, China (CHB)
- Utah residents with ancestry from northern and western Europe (CEU)
A copy number variation (CNV) is a segment of DNA from on kilobase to several megabases in which copy-number differences have been found by comparison of two or more genomes.
CNVs may either be inherited or caused by de novo mutation. They are assumed to be related with diseases like lung cancer, HIV infection, inflammatory autoimmune disorders, autism, schizophrenia, and idiopathic learning disability (see also http://en.wikipedia.org/wiki/Copy_number_variation).
Data preparation:
We first preprocessed the raw data (CEL files) by cn.FARMS
with 100kb windows that contained at least two probe sets locations.
After excluding the X and the Y chromosome, 2548 locations
obtained an INI call (informative call). The cn.FARMS curated
values of these 2548 locations across 269 Hapmap
individual served as input to FABIA.
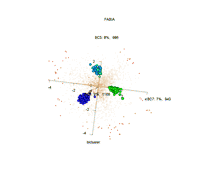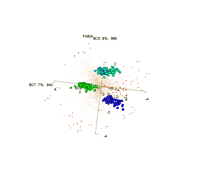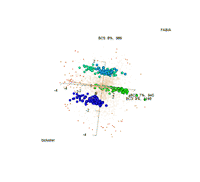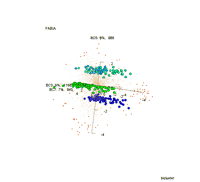
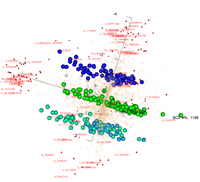
Visualization of 3 biclusters that were found by FABIA:
Publication:
McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, Elliott AL, Parkin M, Hubbell E, Webster T, Mei R, Veitch J, Collins PJ, Handsaker R, Lincoln S, Nizzari M, Blume J, Jones KW, Rava R, Daly MJ, Gabriel SB, Altshuler D.
Integrated detection and population-genetic analysis of SNPs and copy number variation.
Nature Genetics 2008 Oct; 40(10):1166-74.
HapMap data sources:
Phase III data (including phase II data)
http://hapmap.ncbi.nlm.nih.gov/downloads/raw_data/hapmap3_affy6.0/
Phase II data
http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000049.v1.p1
The data from the individual "NA12236" (female, CEU) was missing.