Copy number analysis of German outbreak strain E. Coli EHEC O104:H4 (TY-2482, LB226692) by comparing it to strain K12 DH10B at Ion Torrent sequencing data
Günter Klambauer*, Martin Heusel, Djork-Arne Clevert, Sepp Hochreiter
Institute of Bioinformatics, Johannes Kepler University, Linz, Austria
klambauer@bioinf.jku.at
We estimated the DNA copy numbers of the 2011 German outbreak strain E. coli O104:H4 (TY-2482 and LB226692 from BGI ftp://ftp.genomics.org.cn/pub/Ecoli_TY-2482/) on Ion Torrent sequencing data. The O104:H4 copy numbers are given in relation to the copy numbers of the E. coli strain K12 DH10B (from http://www.edgebio.com/services/iontorrent.php, download instructions at the bottom). As reference genomes, to which the sequencing reads are mapped, we used E. coli strains DH10B, 55989, and O157:H7. Including the sequencing data from K12 strain in our analysis allowed us to separate technical / biological variations from true copy numbers. For copy number estimation, we used our probabilistic model cn.MOPS (http://www.bioinf.jku.at/software/cnmops/cnmops.html) which has been designed for estimating copy numbers in next generation sequencing data. cn.MOPS decomposes read count variations into variation caused by DNA copy numbers and random/noisy variations.
Major results concerning the O104:H4 genome:
Shiga toxin stx2 is amplified (two copies) in O104:H4 (red arrows in Fig. 1)
Tellurite gene cluster ter are present in O104:H4 (green arrows in Fig. 1)
Intimin gene eae is missing in O104:H4 (light blue arrow in Fig. 1)
β-lactamases ampC, ampD, ampE, ampG, and ampH are present in O104:H4
A large copy number 2 region from 610kbp to 705kbp (DH10B as reference) or 675kbp to 770kbp (O157:H7 as reference) has been detected in DH10B
Plasmids that were top ranked by cn.MOPS' call:
plasmid pKL1 (cn.MOPS call of 1.6) containing one gene repA which controls the plasmid copy number
plasmid pO26-S1 (cn.MOPS call of 1.36) found in the Shiga-toxin producing E. coli strain O26
plasmid pEC_Bactec (cn.MOPS call of 1.02) containing genes TEM-1 (common plasmid encoded beta-lactamase) and CTX-M-15 (extended-spectrum beta-lactamase gene)
plasmid pMG828-1 (cn.MOPS call of 0.71) containing one gene encoding a replication protein
plasmid 55989p (cn.MOPS call of 0.71) containing the genes aatA (ABC-transporter protein gene) and aggR (master regulator gene of Vir-plasmid genes)
plasmid pAPEC-O1-R (cn.MOPS call of 0.35) carrying an antibiotic resistance gene cluster
plasmid pO86A1 (cn.MOPS call of 0.2) containing the gene aatA (ABC-transporter protein gene)
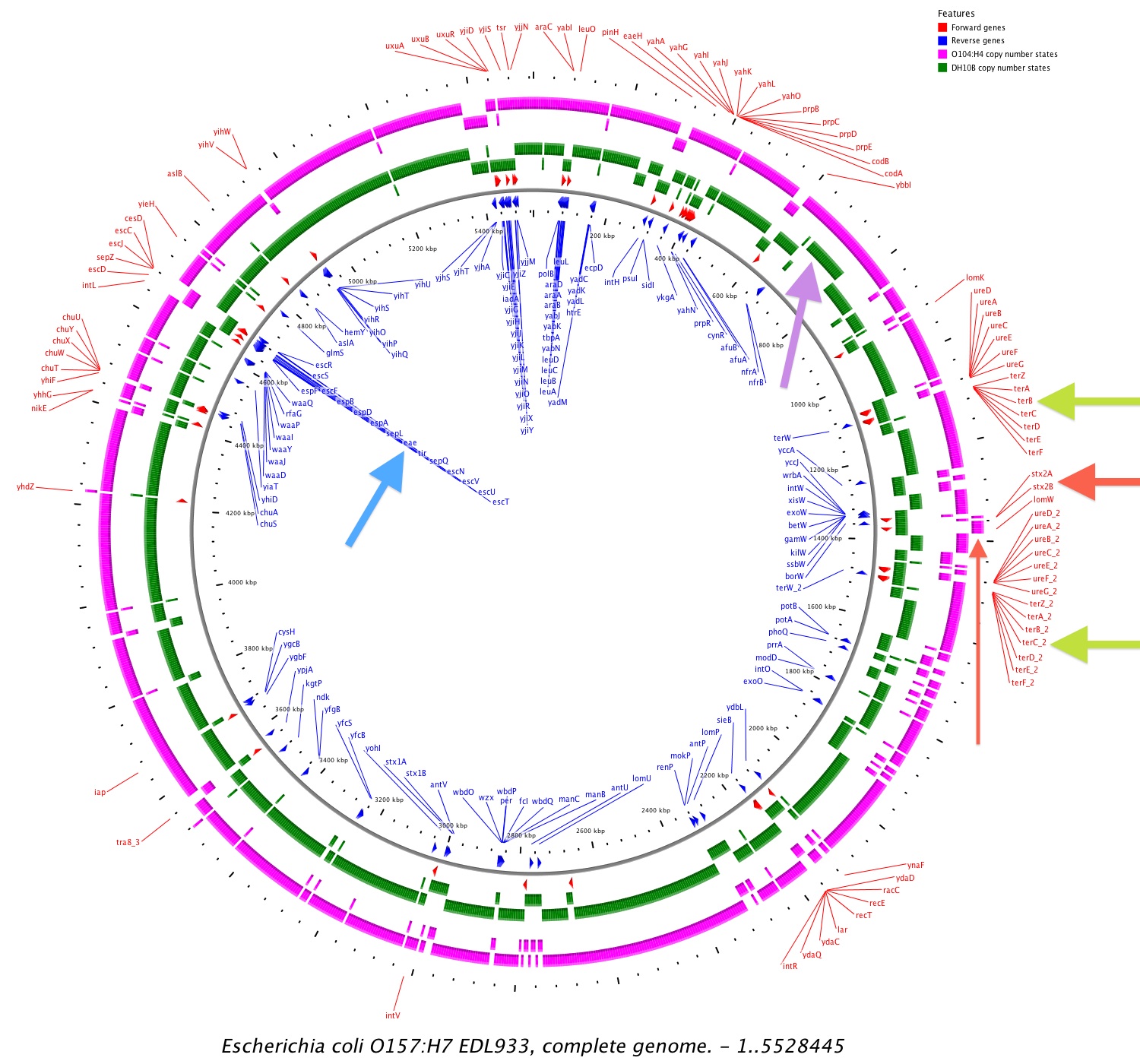
Figure 1. Copy numbers variable regions of O104:H4 compared to DH10B if mapped to O157:H7. Red arrows: shiga toxin stx2 is amplified in O104:H4; Green arrows: tellurite gene cluster ter are present in O104:H4; Light blue arrow: intimin gene eae is missing in O104:H4. Violet arrow: copy number 2 region from 675kbp to 770kbp detected in DH10B. Figure as SVG.
Copy number table of the outbreak strain O104:H4 and DH10B using reference O157:H7:
- tab-delimited file
integerCNTable_ref_O157.txt
- Microsoft Excel file
integerCNTable_ref_O157.xls
- tab-delimited file
integerCNTable_ref_55989.txt
- Microsoft Excel file
integerCNTable_ref_55989.xls
- tab-delimited file
integerCNTable_ref_K12.txt
- Microsoft Excel file
integerCNTable_ref_K12.xls
Mapping and copy number statistics for reference O157:H7:
- Copy number distribution tables, integer copy number plot, ratio plot, read count plot:
cn_O157_H7_AE_LB226692_eps0.05_5kpb_gk.pdf
Mapping and copy number statistics for reference 55989:
- Copy number distribution tables, integer copy number plot, ratio plot, read count plot:
cn_55989_LB226692_eps0.05_5kbp_gk.pdf
Mapping and copy number statistics for reference K12-DH10B:
- Copy number distribution tables, integer copy number plot, ratio plot, read count plot:
cn_K12_LB226692_eps0.05_5kbp_gk.pdf
Discussion of our results
Our results reconfirm findings of the Robert-Koch-Institute (see summary ftp://ftp.genomics.org.cn/pub/Ecoli_TY-2482/2011vs2001_v2.xls) that the outbreak strain E. coli O104:H4 is:
shigatoxin 2 (stx2) positive
intimin negative
The
shiga toxin stx2 is characteristic for O104:H4 as a manuscript of
the "Bundesinstitut für Risikobewertung" states
(http://www.bfr.bund.de/cm/349/enterohaemorrhagic_escherichia_coli_o104_h4.pdf).
That
shiga toxin is produced by O104:H4 was reported in
a NatureNews article
(http://www.nature.com/news/2011/110609/full/news.2011.360.html)
from June 9th by Marian
Turner who writes “The bacterium in this outbreak, currently
recognised as strain O104:H4, makes Shiga toxin, which is responsible
for the severe diarrhoea and kidney damage in patients whose E. coli
infections develop into haemolytic uremic syndrome (HUS).”
Our
finding that tellurium resistance genes are present in O104:H4
has also been reported in NatureNews
(http://www.nature.com/news/2011/110602/full/news.2011.345.html)
at June 2nd by Marian
Turner who writes “In addition to the antibiotic-resistance genes,
the bacteria contain a gene for resistance to the mineral tellurite
(tellurium dioxide)”.
In the same article
Marian Turner writes “One telltale sign is that the strain does not
contain the eae gene, which codes for a protein called intimin, an
adhesion protein that allows the bacteria to attach to cells in the
gut” which we also confirmed. Our findings concerning
β-lactamases are of interest as they tend to make bacteria resistant
to common antibiotics.
For
additional information and analysis see: Kat Holt’s
analysis in
http://bacpathgenomics.wordpress.com/2011/06/07/stecehec-outbreak-horizontally-transferred-genes/
Manuscript in
Nature precedings “Escherichia coli EHEC Germany outbreak
preliminary functional annotation using BG7 system”, Marina
Manrique, Pablo Pareja-Tobes, Eduardo Pareja-Tobes, Eduardo Pareja,
and Raquel Tobes
(http://precedings.nature.com/documents/6001/version/1).
GitHub Wiki
https://github.com/ehec-outbreak-crowdsourced/BGI-data-analysis/wiki/:
“In this wiki we aim to gather all the results of the analysis of
the E. coli O104:H4 strain responsible for the current outbreak in
Germany and Europe.”