DeepTox: Deep Learning for Toxicity Prediction
DeepTox is a pipeline for predicting toxic effects of chemical compounds.
The Tox21 Data Challenge has been the largest effort of the scientific community to compare computational methods for toxicity prediction. This challenge comprised 12,000 environmental chemicals and drugs which were measured for 12 different toxic effects by specifically designed assays. We participated in this challenge to assess the performance of Deep Learning in computational toxicity prediction. Deep Learning has already revolutionized image processing, speech recognition, and language understanding but has not yet been applied to computational toxicity. Deep Learning is founded on novel algorithms and architectures for artificial neural networks together with the recent availability of very fast computers and massive datasets. It discovers multiple levels of distributed representations of the input, with higher levels representing more abstract concepts. We hypothesized that the construction of a hierarchy of chemical features gives Deep Learning the edge over other toxicity prediction methods. Furthermore, Deep Learning naturally enables multi-task learning, that is, learning of all toxic effects in one neural network and thereby learning of highly informative chemical features. In order to utilize Deep Learning for toxicity prediction, we have developed the DeepTox pipeline. First, DeepTox normalizes the chemical representations of the compounds. Then it computes a large number of chemical descriptors that are used as input to machine learning methods. In its next step, DeepTox trains models, evaluates them, and combines the best of them to ensembles. Finally, DeepTox predicts the toxicity of new compounds. In the Tox21 Data Challenge, DeepTox had the highest performance of all computational methods winning the grand challenge, the nuclear receptor panel, the stress response panel, and six single assays (teams .Bioinf@JKU.). We found that Deep Learning excelled in toxicity prediction and outperformed many other computational approaches like naive Bayes, support vector machines, and random forests.
-
DeepTox was the best performing method in the
Tox 21 Data Challenge
where it won the Grand Challenge, Stress Response Panel, Nuclear Receptor Panel and six of twelve subchallenges.
For detailed results see here.
- DeepTox was the best performing method in the NIEHS-NCATS-UNC DREAM Toxicogenetics Challenge 2013 at predicting the average cytotoxicity of 50 compounds (Team "Austria").
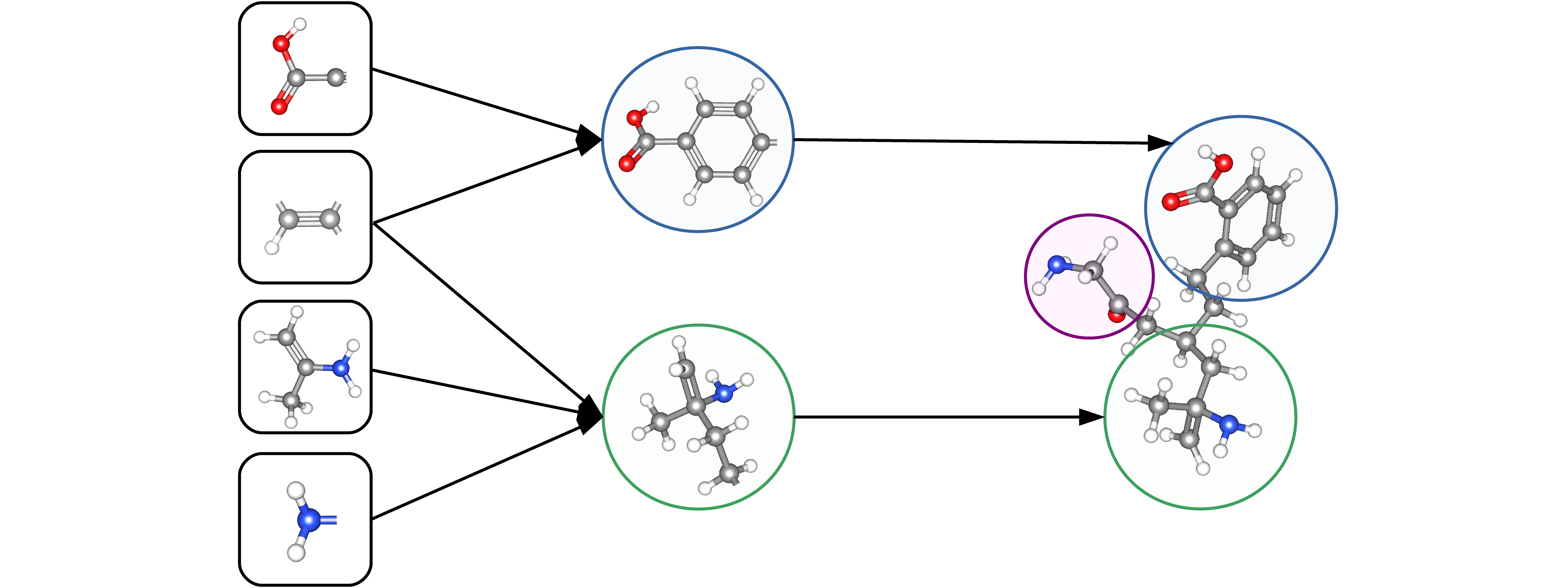 Neural networks constructing complex features, such as pharmacophores, out of simpler substructures. |
Publication
-
Published Article (Frontiers in Environmental Science)
- The Supplementary material can be obtained from the Frontiers page of the article.
Dataset
A preprocessed Tox21 Dataset is available at http://bioinf.jku.at/research/DeepTox/tox21.html.Citation
Mayr Andreas, Klambauer Günter, Unterthiner Thomas, Hochreiter Sepp (2016). DeepTox: Toxicity Prediction using Deep Learning. Frontiers in Environmental Science. 3(80)BibTeX:
@article{DeepTox,
title={{DeepTox: Toxicity Prediction using Deep Learning}},
author={Mayr, Andreas and Klambauer, G{\"u}nter and Unterthiner, Thomas and Hochreiter, Sepp},
journal={Frontiers in Environmental Science},
volume={3},
year={2016},
number={80}
}
Architecture of the pipeline
The following methods are the crucial parts of DeepTox:
- Deep Learning: Relying on our long expertise in the field of neural networks we used Deep Learning as a major method for toxicity prediction
- Kernel-based Structural and Pharmacological Analoging (KSPA): A new method that improves predictions by exploiting information that is available in public databases.
Deep Learning was implemented on nVidia GPUs. The software is available at GitHub.
The following methods are also part of DeepTox:
- Machine Learning methods: SVMs with various kernels, Random Forests, Elastic Nets.
- Features and kernels: ECFP; DFS; 3D features based on MOPAC; Quantum-mechanical descriptors; Tanimoto, Minmax and various 2D, 3D and pharmacophore kernels; in-house toxicophore and scaffold features.
Acknowledgements
The authors gratefully acknowledge the support of NVIDIA Corporation with the donation of a GPU used for this research.
Further References
Hochreiter, S. and Schmidhuber, J. (1997a). Flat minima. Neural computation 9(1):1–42.
Hochreiter, S. and Schmidhuber, J. (1997b). Long short-term memory. Neural computation 9(8):1735–1780.
Hochreiter, S. (1998). The vanishing gradient problem during learning recurrent neural nets and problem solutions. International Journal of Uncertainty, Fuzziness and Knowledge-Based Systems 6(02):107–116.36.
Hochreiter, S., & Schmidhuber, J. (1999). Feature extraction through LOCOCODE. Neural Computation 11(3): 679-714.
Hochreiter, S., Younger, A. S., & Conwell, P. R. (2001). Learning to learn using gradient descent. In Artificial Neural Networks—ICANN 2001 (pp. 87-94). Springer Berlin Heidelberg.
Unterthiner, T.; Mayr, A.; Klambauer, G.; Steijaert, M.; Ceulemans, H.; Wegner, J. K.; & Hochreiter, S. (2014). Deep Learning as an Opportunity in Virtual Screening. Workshop on Deep Learning and Representation Learning (NIPS2014).
Unterthiner, T.; Mayr, A.; Klambauer, G.; & Hochreiter, S. (2015). Toxicity Prediction using Deep Learning. CoRR, abs/1502.02072, 2015.
Günter Klambauer, Bie Verbist, Liesbet Vervoort, Willem Talloen, QSTAR Consortium, Ziv Shkedy, Olivier Thas, Andreas Bender, Hinrich W.H. Göhlmann, Sepp Hochreiter (2015). Using transcriptomics to guide lead optimization in drug discovery projects, Drug Discovery Today, Available online 10 January 2015, ISSN 1359-6446, (http://dx.doi.org/10.1016/j.drudis.2014.12.014.)
Günter Klambauer, Michael Mahr, and Sepp Hochreiter (2013) Rchemcpp: An R package for computing the similarity of molecules. Rchemcpp at Institute of Bioinformatics, Web-service for structural analoging, Rchemcpp Package at Bioconductor.org

